Bacterial or viral: To treat or not to treat with antibiotics
04 Sep 2017
Written by Eran Eden, Kfir Oved, Tanya Gottlieb
Episode I – The global challenge
Many are familiar with this ‘seemingly’ simple scenario that occurs many times a day across the developed and developing world. Someone, maybe it is yourself, your child or an elderly relative, visits the doctor with a fever. Commonly, assigning a diagnosis comes down to deciding whether the infection is bacterial or viral. Depending on the outcome we may or may not treat with antibiotics, but… how do we decide?
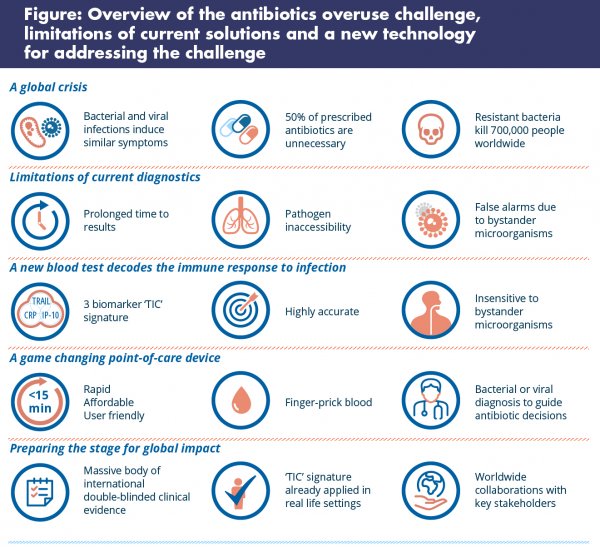
Antibiotics do not work on viral infections. Unfortunately, though, bacterial and viral infections often present with very similar symptoms, causing uncertainty that leads to antibiotics being used, in many instances, when they are not needed. This antibiotic misuse contributes to the rise of antimicrobial resistance.
In the 21st century, when we can sequence genomes and take soil samples from Mars, why is this seemingly simple decision about antibiotics still so challenging? The reason is that even in resource-rich settings where cutting edge microbiological tools are available, and certainly in resource limited settings, actionable diagnosis is still constrained by:
- The prolonged waiting time for results, which can be hours or days;
- Infections inside the body that are not easy to sample, which occur quite often, and so are difficult to diagnose by direct pathogen detection, for instance, pneumonia;
- Inability to interpret whether detected microbes are actually causing the disease or a mere bystander.
Over 10 years ago, we decided to take on the challenge of developing a new tool that can help clinicians decide if an infection is bacterial or viral, and founded a biotech company, MeMed.
Episode II – Relying on the best detection system: The immune system
We initially ‘played’ with different technologies that detect pathogens, but began wondering whether this approach is really suited to the clinical question of whether one has a bacterial or viral infection. What if we flip the problem upside down? What if rather than trying to directly detect the pathogen, we develop a solution that relies on the best detection system out there – much better than we could ever hope to create as scientists, clinicians and engineers – our own immune system?
We decided to take on the challenge of developing a new tool that can help clinicians decide if an infection is bacterial or viral.
We began searching for ways to decode how the immune system responds differently to bacterial as opposed to viral infections.
Using the individual’s immune system is not only a nice twist to the story, but provides the solution to some of the constraints of classical technologies, as this approach can lead to:
- Rapid results, even within minutes (particularly in the case of circulating host-proteins that unlike other types of biomarkers, allow very rapid, quantitative and affordable measurements);
- Diagnosis even when the infection site is inaccessible, because the immune system circulates throughout the body;
- Prevention of false alarms due to colonisation, as the immune system is primarily triggered by disease-causing pathogens, and is not fooled by bystanders.
To turn the idea into reality, we joined forces with collaborators from across the globe.
Episode III – ‘No magic bullets’ – it’s all about biomarker team work
There are a few protein biomarkers in the immune system that predominantly increase in response to bacteria, such as C-reactive proteins (CRP) and procalcitonin, which clinicians sometimes monitor to help with infection diagnosis. Although each has merit, they also have blind spots.
We asked ourselves: What if we could discover novel protein biomarkers that increase in response to viral infections? Then maybe, we could identify a unique combination of viral-induced and bacterial-induced biomarkers, where participating proteins cover for one another’s blind spots. Taking this approach, we set out to discover an ‘immune fingerprint’ that could be broadly applicable to help with infection diagnosis.
Finding the key biomarkers and identifying the useful combination took us over four long years. Along the way we saw many ‘faces in the clouds’ – biomarkers that initially appeared promising, but turned out to be artefacts and had to be discarded. After in silico, and in vitro work, we ultimately ran our first multi-centre prospective clinical study recruiting 1,002 adults and pediatric patients. We called it “Curiosity”.
In this study, we quantitatively screened over 600 proteins in patients with bacterial or viral infections (and as far as we know, it was the largest host-protein screening to date!). The best performing combination out of the numerous ones rigorously interrogated, included three protein biomarkers called: TRAIL, IP-10, and CRP. Nicknamed the ‘TIC’ signature, this trilogy of proteins demonstrated robust performance, with sensitivity and specificity over 90% across different ages, pathogens, times from symptom onset – outperforming individual proteins and other routine laboratory parameters.
When the results were finally published in a peer-reviewed journal, an interviewer from BBC made the following comments: “These results are indeed promising and even potentially transformative, but I’m still not convinced. To be persuaded you need to conduct several external double-blind studies, that reproduce your original results, and that takes years. Ah… and usually biomarkers don’t survive this type of stringent double-blind validation.”
She was right. The history of translational sciences has many examples of biomarkers that initially looked promising, but did not pass double-blind evaluation. Little did she know, however, that we had already embarked on this ‘double-blind journey’ two years previously!
Episode IV – Sidestepping the graveyard of new biomarkers
Together with partners from leading medical centers, universities and other organisations we spent four years running two external double blind-clinical studies called ‘Opportunity’ and ‘Pathfinder’ (ntotal=1,374), each independently evaluating the diagnostic performance of the TIC signature. If our initial results had not been externally reproducible it would probably have been game over – but they were reproducible.
Encouraged by these data, we and others have expanded our efforts to further solidify the clinical evidence in various populations in more than ten clinical studies around the world (enrolling more than 10,000 patients), including one with the University College London. These studies are currently ongoing.
Little did she know, however, that we had already embarked on this ‘double-blind journey’ two years previously!
Additionally, we are gearing up to study the applicability of the TIC signature in the developing world, where there is urgent need for reliable tests to aid in the diagnosis of non-malarial febrile illness, particularly tests that take into account different co-morbidities, such as malnutrition. The influence of chronic diseases common to particular regions (e.g., tuberculosis) will need to be carefully examined.
In parallel to clinically validating the TIC signature, we worked on creating a product, as we knew this was necessary to bring the test to patients in the real world.

Episode V – From clinical research to the patient bedside
Meanwhile, our team grew to include experts in emergency medicine and infectious disease, pediatrics, molecular immunology, software and mechanical engineering, big data analytics, analytical chemistry, in vitro diagnostics, industrial design, marketing, health economics, reimbursement and regulation.
Initially, we developed a first generation product based on the TIC signature called ImmunoXpert™ that is laboratory-based and takes two hours, just to launch and learn. Today ImmunoXpert™ (CE-IVD) has been used to diagnose infections in more than 10,000 children and adults as part of our early access program, which is steadily expanding.
Having an accurate and validated product is a must – but we knew from the get-go that it’s not enough. To potentially have a big impact, the test needs to be fast, affordable and easy to use, too.
Episode VI – Preparing the stage for global impact
For global use, one needs a device that can measure the TIC signature in a very rapid (within minutes), affordable and user-friendly manner. We have been working on this next generation product for quite some time and look forward to telling you more about it in the near future.
